Variant Genome Sequencing for Public Health Authorities* and Identifying the Omicron Variant: Conversations with Curative Scientists

6 Minutes

Dr. Mark Kelleher

Feb 12, 2022
SARS-CoV-2 and mutations have resulted in newer, more infectious variants of concern, such as Delta and Omicron. Curative’s research and development division includes the next generation sequencing development group led by Mikhail Hanewich-Hollatz, PhD, which contributes to global and local efforts on the SARS-CoV-2 variants surveillance.
Mikhail, together with scientist Eugenia Khorosheva, PhD, and research associates Sophia Quasem and Janet Chen, develops and operates at the cutting edge of technology to help track the COVID-19 pandemic through identifying variants based on genome sequencing.
Their work is essential in helping to determine public health decisions and overall knowledge of the virus. As new variants emerge and take hold, our team is monitoring and identifying cases and submitting data to public health organizations. Here is what we’ve learned from discussing Curative’s next generation sequencing with Eugenia and Mikhail.


Mikhail Hanewich-Hollatz, PhD, Associate Director and Eugenia Khorosheva, PhD Scientist at Curative Research and Development
Curative decided to invest in a sequencing research laboratory in 2020. Starting the new project on sequencing COVID-19 variants was a highly collaborative venture for Mikhail’s group. They had to organize a new lab space, develop the operating procedures, obtain the tools and automation needed to be at the forefront of innovation, select and test the best biochemical strategies for severe acute respiratory syndrome-2 (SARS) sequencing, and develop needed pipelines for results interpretation and reporting. This was made possible through collaboration with a number of Curative teams, including the purchasing and facilities teams, automation team, positive sample team, clinical trials team, and the NGS R&D group led by Jean Xie, particularly with bioinformatician Julio Cesar Espinoza.
Currently in Curative’s NGS Development lab, SARS-CoV-2 sequencing results vary from approximately 3-4 days to 6-7 days for 376-752 samples. Starting in the spring of 2021, Mikhail, Eugenia, Sophia and Janet have sequenced and reported approximately 1% of all known SARS-CoV2 genomes in California and 0.5% nationally. For a company that started in 2020 and acquired a lab at the end of that year, we are proud to have made such a contribution to public knowledge.


Members of the Curative Research and Development/NGS Development Team, Sophia Quasem and Janet Chen are at work, preparing sequencing libraries for SARS-CoV-2 Next Generation Sequencing
As the pandemic progressed and new variants of SARS-CoV-2 evolved, we contributed to efforts on the local variants surveillance and early detection of “variants of concern”, which the World Health Organization defines as a variant that increases transmissibility or detrimental change to COVID-19 epidemiology, increases in virulence or changes clinical disease presentation, or decreases in the effectiveness of public health and social measures or available diagnostics, vaccines, and therapeutics.
In every SARS-CoV-2 sample, the determined SARS-CoV-2 gene sequences define the shape and function of the coded viral proteins. Therefore, sequencing of the viral genome is thought to be the simplest way to track known any new changes in a particular virus. It helps in predicting the isolate’s infectivity, transmissibility, pathogenicity, as well as its ability to escape host immune system defense or available therapeutics interventions. This information is important to share with other scientists, epidemiologists, public health officials, and even physicians in some cases.
All Curative sequencing data for SARS-CoV-2 isolates are reported and uploaded to a global public repository of SARS-CoV-2 sequences called GISAID, where there are currently over 6,000 individual SARS-CoV-2 genomes searchable by the key word “Curative”. To see Curative’s impact, you can view our catalog of entries in GISAID.
Above is a catalog of Curative’s contributions to GISAID, which is used for sequencing data submission globally and promotes the rapid sharing of data on the coronavirus causing COVID-19.
We found that SARS-CoV-2 variant surveillance could be conducted in a private laboratory in a timely and accurate manner, meaning our facilities were well-equipped to provide critical knowledge at a rapid pace. SARS-CoV-2 positivity rates from specimens tested in a private laboratory were similar to county level data collected during the same periods.
When the variant sequencing data is put on the timescale, we can observe which variants are most common in a state over time, and see early how vaccination in populations eliminates and cuts off branches in viral evolution and leaves only those non-numerous variants that are known to be capable of infecting vaccinated people. Among the samples that tested positive for SARS-CoV-2, we observed that initially, the Alpha variant of SARS-CoV-2 was most prevalent in Los Angeles and Riverside County in May 2021, however, the Delta variant of SARS-CoV-2 became dominant over a period of weeks.
Among the samples identified as the Delta variant, a large number carried mutations classifying them as sub-lineages of Delta, which is evidence of continued mutation. We are continuing to monitor the spread of Delta and we are on the watch as Omicron spreads across the country. We are ready to relay this information to inform public health decisions.
Our work on tracking these changes in LA County is done in collaboration with Lauren Lopez, the Clinical Trials team, and other research and development scientists. Our results are available in our pre-printed manuscript titled Local Monitoring of SARS-CoV-2 Variants in Two Large California Counties in 2021.
CAPTION FOR IMAGES BELOW: On the top is the public tracking of variants over time in GISAID, and the light blue color indicates the emergence of Delta (Fig.1A). On the bottom is Curative’s own data plotted separately from May to September 2021 (Fig. 1B) and from September 2021 to January 2022, which closely resembles that of the national and global trends (Fig.1C).
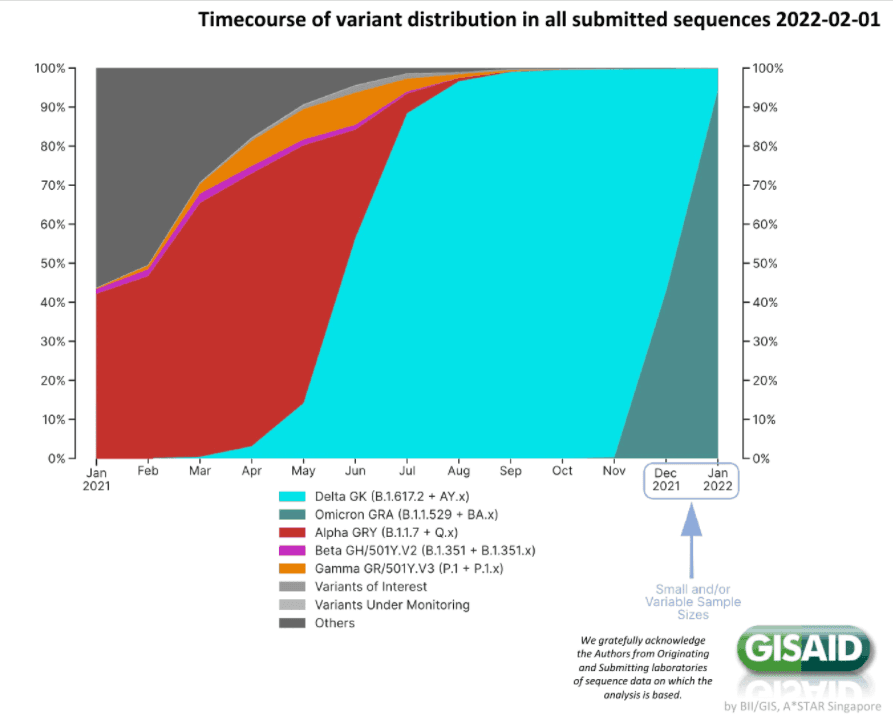
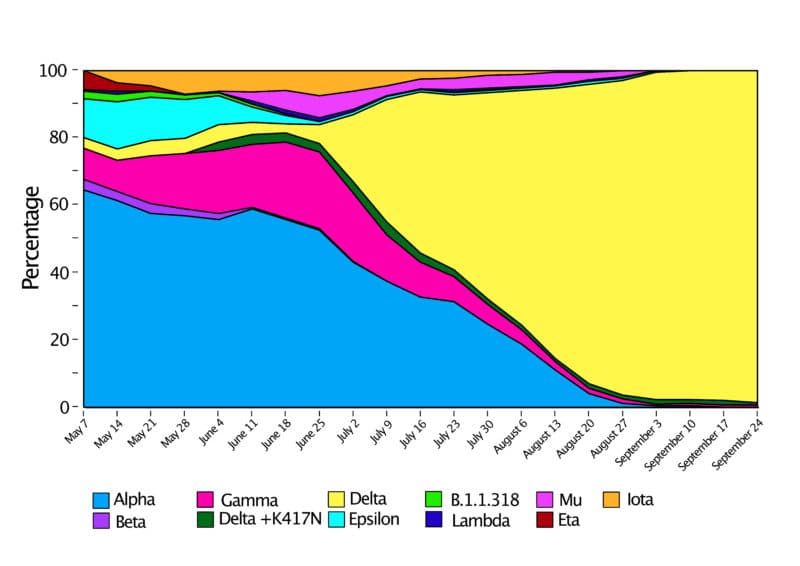
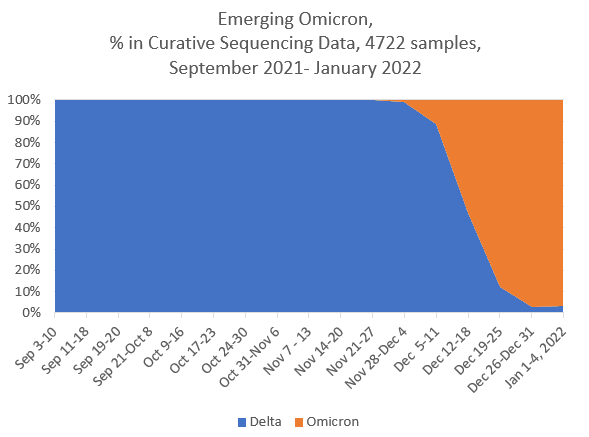
When compared to publicly available data on variants, our sampling found similar proportions of variants of concern in our samples over time. As the COVID-19 pandemic persists, we are anticipating the emergence of new variants.
As the United States focuses on the Omicron variant that is spreading rapidly, increase our sequencing throughput in order to be able to sequence more samples and detect the variant as soon as it appears in any of our positive samples. As we witnessed the Delta variant’s continued mutations, we expect to see similar trends as the Omicron variant spreads nationwide. We plan to relay this information to inform public health decisions as soon as it is available.
We are continuing to invest in innovation that demonstrates our commitment to public health, and our next generation sequencing development team is a key part of that initiative. In 2022, the team is aiming to expand sequencing capabilities by at least four times. This is happening with the support of Curative Senior Vice President Mark Kelleher and Vice President Yuriy Shevchenko, and many teams throughout the company.
To learn about our work, take a deep dive into our Monrovia lab in our article, “Curative is Positioned to Take on Delta and Any Future Variants — the Monrovia Lab with Jean Xie”.
*Public Health Authorities are legally authorized to receive reports of variant sequencing data under 45 CFR 164.512(b)(1)(i)
Sign up for our Newsletter
Table of Contents








